TidyMass an object-oriented reproducible analysis framework for LC–MS data
Abstract
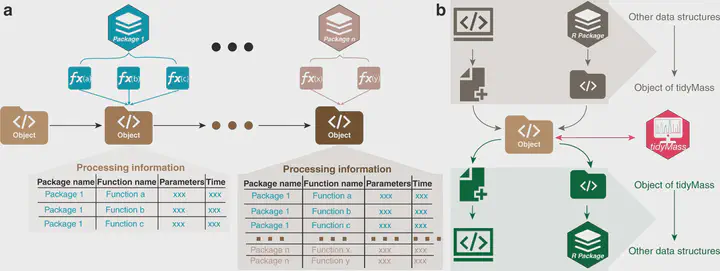
Reproducibility, traceability, and transparency have been long-standing issues for metabolomics data analysis. Multiple tools have been developed, but limitations still exist. Here, we present the tidyMass project (https://www.tidymass.org/), a comprehensive R-based computational framework that can achieve the traceable, shareable, and reproducible workflow needs of data processing and analysis for LC-MS-based untargeted metabolomics. TidyMass is an ecosystem of R packages that share an underlying design philosophy, grammar, and data structure, which provides a comprehensive, reproducible, and object-oriented computational framework. The modular architecture makes tidyMass a highly flexible and extensible tool, which other users can improve and integrate with other tools to customize their own pipeline.